Research Article
RAF Kinase Inhibitory Protein Expression and Phosphorylation Profiles in Oral Cancers
Hallums DP1, Gomez R1, Doyle AP2, Viet CT4,5, Schmidt BL4,5 and Jeske NA1,2,3*
1Departments of Oral and Maxillofacial Surgery, University of Texas Health Science Center at San Antonio, USA
2Departments of Pharmacology, University of Texas Health Science Center at San Antonio, USA
3Departments of Physiology, University of Texas Health Science Center at San Antonio, USA
4Department of Oral Maxillofacial Surgery, New York University, USA
5Department of Oral Maxillofacial Surgery, Bluestone Center for Clinical Research, New York University, USA
*Corresponding author: Nathaniel A. Jeske, Department of Oral & Maxillofacial Surgery, University of Texas Health Science Center of San Antonio, Center for Biomedical Neuroscience, 7703 Floyd Curl Dr., San Antonio, TX 78229-3900, USA
Published: 01 Sep, 2016
Cite this article as: Hallums DP, Gomez R, Doyle AP, Viet CT, Schmidt BL, Jeske NA. RAF Kinase Inhibitory Protein Expression and Phosphorylation Profiles in Oral Cancers. Clin Surg. 2016; 1: 1100.
Abstract
Raf Kinase Inhibitory Protein (RKIP) expression has been profiled for a number of unique tissue
cancers. However, certain tissues have not been explored, and oral and oropharyngeal cancers stand
out as high priority targets, given their relatively high incidence, high morbidity rate, and in many
cases, preventable nature. The purpose of this study was to examine changes in RKIP expression
and phosphorylation in tissues resected from oral cancer patients, and compare to results generated
from immortalized cell lines raised from primary oral cancer tissues, including oral squamous cell
carcinoma line 4 (SCC4) and human squamous cell carcinoma line 3 (HSC3). Out of 4 human
samples collected from male and female patients across various ages with variable risk factors, we
observed an across the board reduction in RKIP expression. Two human samples demonstrated a
significant increase in phosphorylated RKIP when normalized to total RKIP, however all 4 were
increased when normalized to total cellular protein. The immortalized oral cancer cell culture HSC3
revealed significant increases in phosphorylated RKIP with no change in total RKIP expression, while
line SCC4 demonstrated an increase in both total and phosphorylated RKIP. Results presented here
indicate that oral cancers behave similarly to other cancers in terms of changes in RKIP expression
and phosphorylation, although immortalized cell line expression profiles significantly differ from
human tissue biopsies.
Keywords: Oral cancers; Raf kinase; Phosphorylation
Introduction
Oral squamous cell carcinoma is the most common cancer in the orofacial cavity, accounting
for approximately 90% of all cancers of the mouth [1]. The American cancer society estimates that
approximately 37,000 people will be diagnosed with oral or oropharyngeal cancer in 2014, and an
estimated 7,300 people will die. These values represent 2.22% of the American population, which is
greater than many other cancer types. However, little proteomic profiling has been conducted on
oral cancers, leaving many clinical researchers with few potential targets for therapeutic treatment.
In this report, we tracked the expression profile for Raf Kinase Inhibitory Protein (RKIP, also known
as phosphatidylethanolamine-binding protein or PEBP) in oral cancer cells.
RKIP is a scaffolding protein capable of binding to and inhibiting Raf kinase [2]. Raf kinase
classically participates in the Ras/Raf/MEK/ERK kinase cascade that transfers mitogenic signals
from the cell membrane to the nucleus [3]. Therefore, RKIP has the capacity to interrupt cell
differentiation and growth, depending on the cell signal being received. In its monomeric form,
RKIP binds to Raf-1 kinase, preventing downstream signal transduction, thereby maintaining
transcriptional events at basal levels. However, upon cell stimulation, phosphorylation of RKIP at
Ser153 results in dimerization of the scaffolding protein, causing it to switch molecular attractions
from Raf-1 to G-protein Receptor Kinase 2 (GRK-2) [4]. This switch can allow Raf-1 to signal
downstream in an unrestricted fashion, resulting in cellular transcription, differentiation, and
growth. Additional RKIP scaffolding to NFκB, which normally mediates cellular apoptosis, can also
be interrupted by phosphorylation, resulting in anti-apoptotic signaling mechanisms that preserve
and prolong cellular integrity [5]. The participation of RKIP in these ubiquitous signaling pathways
highlights the importance of this protein to cancer formation and metastasis.
Previous studies identify RKIP as an important protein
determinant in many types of cancers, including prostate, melanoma,
colorectal, liver, and breast [6-10]. Indeed, studies report significant
down-regulation of RKIP expression in differentiated gastric cancer
cells [11] and a number of solid tumors including prostate [7], breast
[12], colorectal [13], and melanoma [14]. Given its role in multiple
signaling events that control significant factors of cell survival, downregulated
RKIP expression would have dramatic results on cellular
growth and phenotype. Further, RKIP phosphorylation, caused
by PKC activation, is associated with poor outcomes in certain
cancers including colon [15]. Therefore, we sought to identify RKIP
phosphorylation and expression profiles in tissue biopsies resected
from oral cancer patients, and compare results to those from widely
used oral cancer cell lines derived from squamous cell carcinomas.
Materials and Methods
Cell/Animal/Patient samples
Oral squamous cell carcinoma line 4 (SCC4) and human
squamous cell carcinoma line 3 (HSC3) cells were grown as previously
described [16]. Cell cultures were left untreated, and in the presence
of 2% serum for 24 hours prior to harvesting for protein extraction.
All procedures utilizing animals were approved by the Institutional
Animal Care and Use Committee of the University of Texas Health
Science Center at San Antonio and were conducted in accordance
with policies for the ethical treatment of animals established by the
National Institute for Health. Male Sprague-Dawley rats 175 – 200g
in weight (Charles River Laboratories, Wilmington, MA) was used
for Trigeminal Ganglia (TG) dissection, as described previously [17].
The study was approved by the Institutional Review Board of New
York University College of Dentistry and University of California San
Francisco (UCSF). All patients provided written informed consent in
accordance with the Declaration of Helsinki. Patients were enrolled
with the following inclusion criteria: 1) biopsy-proven HNSCC,
and 2) no history of prior surgical, chemotherapeutic, or radiation
treatment for oral SCC. Demographic information was collected for
each patient including age, sex, ethnicity, oral SCC location (tongue,
floor of mouth, buccal mucosa, gingival, palate), tumor size (greatest
dimension based on clinical examination), and evidence of metastasis.
At the time of surgical resection a 5x5 mm piece of oral cancer was
collected. A normal piece of oral mucosa measuring 5 x 5 mm was
collected from a contralateral, anatomically matched site.
Western blotting
Cells and tissues were homogenized by 20 strokes in a PotterElvejm
homogenizer on ice in homogenization buffer (25mM
HEPES, 25mM sucrose, 1.5mM MgCl2
, 50mM NaCl, pH to 7.2)
with protease and peptidase inhibitors (aprotinin10µg/ml, leupeptin
10µM, pepstatin 1µg/ml, phenylmethylsulfonyl fluoride (PMSF,
10µg/ml), sodium orthovanadate 100µg/ml) added immediately prior
to harvesting. Homogenates were incubated on ice 15 min, then lysed
with 1% Triton X-100 via 20 passes through a 25g tuberculin needle,
and incubated on ice 10 min. Samples were centrifuged at 1000g
for 1 min at RT to precipitate un-lysed cells, and protein content
was determined by Bradford analysis [Bradford, 1976 #301]. 25 µg
aliquots were resolved by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to polyvinyl difluoride
(PVDF, EMD Millipore, Billerica, MA) membranes for Western
blotting, using antibodies specific to phosphorylated RKIP (Ser 153,
SantaCruz Biotechnology, Santa Cruz, CA), total RKIP (SantaCruz
Biotechnology) and β-actin (SantaCruz Biotechnology). Appropriate
secondary antibodies (GE Healthcare Life Sciences, Piscataway, NJ)
were utilized with enhanced chemiluminescence (GE Healthcare
Life Sciences) to visualize protein immunoreactivity. Samples were
independently analyzed 3-4 times; images are representative of all
trials. Pixel densities of scanned Western blots were quantified using
NIH image 1.62, with samples normalized as indicated in figures.
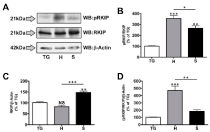
Figure 1
Figure 1
RKIP phosphorylation and expression in oral cancer cell lines.
Primary trigeminal neuron cultures (TG), HSC3 (H) and SCC4 (S) cell lines
were lysed and prepared for proteomic analysis by Western blot. A.Proteins
were identified using antibodies specific for phospho Ser153 RKIP (pRKIP),
total RKIP (RKIP) and β-Actin. Protein molecular weights are shown to the
left of the lots. Densitometry was performed on immunoreactive bands, with
pRKIP normalized to total RKIP (B), total RKIP normalized to β-Actin (C) and
pRKIP normalized to RKIP/β-Actin (D). Results shown are representative of 3
independent trials; statistics determined by one-way ANOVA with Bonferroni
post-hoc correction.
Results
RKIP expression profile in oral cancer cell lines
Oral cancers are widely studied for their invasive morbidity,
yet little has been reported on expression profiles of transcriptional
regulators. RKIP, an important modulator of the Ras/Raf/MEK/ERK
kinase cascade, has been characterized in multiple cancer types, except
for oral. To begin our investigation, we probed for phosphorylated
and total RKIP expression in immortalized oral cancer cells lines
oral squamous cell carcinoma line 4 (SCC4) and human squamous
cell carcinoma line 3 (HSC3). Cell lysates were generated of these
cells under naïve, low serum conditions, to more closely represent
physiological significance. Furthermore, we compared the protein
expression profiles of these cancer cell lines to that of a primary
cell model, Trigeminal Ganglia (TG) neurons. The inclusion of other
immortalized cell lines for control purposes would have confounded
result interpretation given that most cell lines are derived from
cancer tissues, and that any immortalized cell line would represent an
abnormal transcriptional environment. In (Figure 1), Western blot
analysis was performed on protein samples taken from cell lysates
of cultured TG neurons, HSC3 cells, and SCC4 cells. Blots were
probed for phosphorylated RKIP (pRKIP, Ser 153), total RKIP, and
β-actin (loading control), and analyzed for densitometry to quantify
expression between cell lines. Firstly, we observed a significant
increase in phosphorylated RKIP when normalized to total RKIP
in both cell lines (Figure 1A-B). Additionally, we observed a 50%
increase in total RKIP in SCC4 cells over the TG control, while
HSC3 cells reveal little change in expression (Figure 1A and 1C).
We then conducted further normalization to account for changes
in RKIP expression (normalized to β-actin) when measuring the
significance of RKIP phosphorylation in the HSC3 and SCC4 cell
lines. As demonstrated in (Figure 1D), HSC3 cells displayed a 5–fold
increase in phosphorylated RKIP when compared to our control TG
cells, while SCC4 cells displayed less than a 1-fold increase. Together,
these data indicate differential RKIP phosphorylation and expression
profiles between commonly used oral cancer cell lines.
RKIP expression profile in oral cancer tissue biopsies
We next performed similar analysis on tissue biopsies of oral
cancers collected from patients. In this analysis, we compared
changes in RKIP expression and phosphorylation between cancerous
biopsies and healthy tissue control samples taken from the same
patient. Therefore, for each patient, normal tissue (N) was compared
to cancerous tissue (C). In (Figure 2), Western blot analysis was
performed on lysates generated from tissue homogenization, and
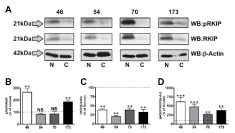
probed for in a similar fashion as in Figure 1. Importantly, we observed
differential RKIP phosphorylation profiles between patient biopsies,
demonstrating significant increases (approximately 250% of control
tissue (sample 46) and 200% (sample 173), (Figure 2A and 2B) in two
patients. Furthermore, all samples produced significantly reduced
total RKIP expression profiles (Figure 2A and 2C). As in Figure 1,
we normalized phosphorylated RKIP measurements to those of RKIP
normalized to β-actin, to accurately identify changes in modified
RKIP while accounting for changes in total RKIP expression. In
(Figure 2D), we observed a significant increase in pRKIP/RKIP/β-
actin for all patient samples over control tissues, indicating that
despite reductions in total RKIP expression, the phosphorylated state
of the protein is significantly higher.
Patient data were examined to determine whether patient history
and/or tumor characteristics could account for sample-to-sample
differences observed in (Figure 2). (Table 1) outlines de-identified
information on each of the patients that cancerous and contralateral
normal tissue samples were taken from. These data sets include, age,
sex, Tumor/Node/Metastasis (TNM) stage, tumor site, and patient
tobacco history. While the buccal mucosa SCC cancer biopsy (P46)
provided the highest pRKIP/RKIP/β-Actin expression profile, there
was little else in the way of correlative descriptive patient data that
could explain the remaining differential RKIP expression profiles.
Taken together, these data support a strong case for reduced RKIP
expression and increased phosphorylated RKIP in human cancer
tissue biopsies. Collectively, these data identify an important
expression profile that is mirrored by many other cancers.
Figure 2
Figure 2
RKIP phosphorylation and expression in oral cancer tissue
biopsies. Tissue biopsies from patient oral cancers (46, 54, 70, and 173) and
matched non-cancerous controls were homogenized, lysed and prepared
for proteomic analysis by Western blot. A. Proteins were identified using
antibodies specific for phosphorylated Ser153 RKIP (pRKIP), total RKIP
(RKIP) and β-Actin. Protein molecular weights are shown to the left of the
lots. Densitometry was performed on immunoreactive bands, with pRKIP
normalized to total RKIP (B), total RKIP normalized to β-Actin (C) and pRKIP
normalized to RKIP/β-Actin (D). Dashed line denotes control expression, set
at 100%. Results shown are representative of 4 independent trials; statistics
determined by one-way ANOVA with Bonferroni post-hoc correction.
Discussion
RKIP transcript and protein expression has been characterized in
numerous cancer types, except for oral cancers. Since oral squamous
cell transformation rate is relatively high compared to other cancers,
changes to mitogenic signaling modulators such as RKIP are highly
significant to understanding the pathogenesis of the disease. Here,
we performed proteomic analysis of RKIP phosphorylation and
expression from two well-studied immortalized oral cancer cell lines
and multiple squamous cell carcinoma biopsies. Our results indicate
that RKIP expression is reduced in cancerous tissue biopsies, similar
to what is found in other cancer types [7,11-14]. Furthermore, we
observed increased RKIP phosphorylation in tissue biopsies, which
parallel observations in other cancer models, and is deemed a
determinant of poor chemotherapeutic prognosis [15]. Interestingly,
only one of the immortalized cell lines (HSC3) provided similar
metrics of RKIP expression and phosphorylation, despite both in vitro
cell lines being used commonly as oral cancer models. Importantly,
HSC3 cells provide a unique in vitro model that mirrors the RKIP
phosphorylation and expression profile characterized in human oral
cancer tissues.
The role of RKIP in cancer and tumor progression was first
identified in prostate cancer cells in 2003 [7]. Here, investigators
identified low RKIP expression in LNCaP prostate cancer cell lines,
and were able to reduce in vitro invasive ability by over-expressing
RKIP. In vivo, increased RKIP expression in implanted C4-2B
cells reduced spontaneous metastasis, but did not affect primary
tumor growth rate, initially identifying RKIP as a metastasis tumor
suppressor gene. This conclusion has been reproduced in several other
cancer types, including breast and urinary bladder [18-20]. Therefore,
RKIP expression may serve as an important prognosticator of
metastatic potential of oral cancers, and can be easily determined by
proteomic means as demonstrated here. Indeed, proteomic analyses
are used currently to detect cell surface markers in breast cancer [21],
whole cell proteomes in prostate cancer [22], and transcription factor
expression profiles for melanomas [23]. Taken together, differential
RKIP characterization of cancerous oral tissues could provide unique
insight into their metastatic potential.
RKIP phosphorylation primarily serves to redirect the protein
away from its inhibition of Raf Kinase and into a scaffolding role
with other signaling proteins, such as G-protein receptor kinase 2
[4,24]. It is interesting to note that we observed a significant increase
in RKIP phosphorylation in all cell and tissue models, although
normalized significance was only present in the HSC3 cell line and
in each of the tissue biopsies. Increased RKIP phosphorylation would
allow for increased Raf signaling downstream to multiple mitogenic
transcription factors, which can stimulate cell growth and division,
and also suppress normal RKIP expression through activation of a
Snail zinc-finger transcription repressor [25]. Therefore, one could
argue that phosphorylation of RKIP by PKC-signaling transduction
contributes to the initial step towards tissue metastasis by effectively
reducing overall RKIP expression via transcriptional repression.
From results presented here, proteomic analysis of oral cancers
could serve as an important screening tool to predict metastatic
potential and assist with determining optimal treatment for the
patient.
Table 1
Table 1
Oral Cancer Patient and Tumor Diagnoses. Vital records were kept for the oral cancer patients (P46, P54, P70, and P173), including age, sex, tumor/nodal/
metastasis (TNM) stage, tumor site, and tobacco history.
Acknowledgement
We acknowledge Cara Gonzales (UTHSCA) for gifting the HSC3 and SCC4 oral cancer cell lines. This work was supported by NIH (NS082746 to NAJ and DE019796 to BLS).
References
- Perez-Sayans M, Somoza-Martin JM, Barros-Angueira F, Reboiras-Lopez MD, Gandara Rey JM, Garcia-Garcia A. Genetic and molecular alterations associated with oral squamous cell cancer (Review). Oncology reports. 2009; 22: 1277-1282.
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999; 401: 173-177.
- Ferrell JE, Jr. MAP kinases in mitogenesis and development. Current topics in developmental biology. 1996; 33: 1-60.
- Lorenz K, Lohse MJ, Quitterer U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature. 2003; 426: 574-579.
- YYeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, et al. Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mole Cellul Biol. 2001; 21: 7207-7217.
- Al-Mulla F, Hagan S, Al-Ali W, Jacob SP, Behbehani AI, Bitar MS, et al. Raf kinase inhibitor protein: mechanism of loss of expression and association with genomic instability. J Clin Pathol. 2008; 61: 524-529.
- Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003; 95: 878-889.
- Li HZ, Gao Y, Zhao XL, Liu YX, Sun BC, Yang J, et al. Effects of raf kinase inhibitor protein expression on metastasis and progression of human breast cancer. Mol Cancer Res. 2009; 7: 832-840.
- Lin K, Baritaki S, Militello L, Malaponte G, Bevelacqua Y, Bonavida B. The Role of B-RAF Mutations in Melanoma and the Induction of EMT via Dysregulation of the NF-kappaB/Snail/RKIP/PTEN Circuit. Genes & cancer. 2010; 1: 409-420.
- Xu YF, Yi Y, Qiu SJ, Gao Q, Li YW, Dai CX, et al. PEBP1 downregulation is associated to poor prognosis in HCC related to hepatitis B infection. J Hepatol. 2010; 53: 872-879.
- . Jia B, Liu H, Kong Q, Li B. RKIP expression associated with gastric cancer cell invasion and metastasis. Tumour Biol. 2012; 33: 919-925.
- Hagan S, Al-Mulla F, Mallon E, Oien K, Ferrier R, Gusterson B, et al. Reduction of Raf-1 kinase inhibitor protein expression correlates with breast cancer metastasis. Clin Cancer Res. 2005; 11: 7392-7397.
- Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ, et al. Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol. 2006; 24: 5672-5679.
- Schuierer MM, Bataille F, Hagan S, Kolch W, Bosserhoff AK. Reduction in Raf kinase inhibitor protein expression is associated with increased Ras-extracellular signal-regulated kinase signaling in melanoma cell lines. Cancer Res. 2004; 64: 5186-5192.
- Cross-Knorr S, Lu S, Perez K, Guevara S, Brilliant K, Pisano C, et al. RKIP phosphorylation and STAT3 activation is inhibited by oxaliplatin and camptothecin and are associated with poor prognosis in stage II colon cancer patients. BMC cancer. 2013; 13: 463.
- Gonzales CB, Kirma NB, De La Chapa JJ, Chen R, Henry MA, Luo S, et al. Vanilloids induce oral cancer apoptosis independent of TRPV1. Oral Oncol. 2014; 50: 437-447.
- Jeske NA, Berg KA, Cousins JC, Ferro ES, Clarke WP, Glucksman MJ, et al. Modulation of bradykinin signaling by EP24.15 and EP24.16 in cultured trigeminal ganglia. J Neurochem. 2006; 97: 13-21.
- Chatterjee D, Bai Y, Wang Z, Beach S, Mott S, Roy R, et al. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J Biol Chem. 2004; 279: 17515-17523.
- Zaravinos A, Chatziioannou M, Lambrou GI, Boulalas I, Delakas D, Spandidos DA. Implication of RAF and RKIP genes in urinary bladder cancer. Pathology oncology research: POR. 2011; 17: 181-190.
- Zhang L, Fu Z, Binkley C, Giordano T, Burant CF, Logsdon CD, et al. Raf kinase inhibitory protein inhibits beta-cell proliferation. Surgery. 2004; 136: 708-715.
- Leth-Larsen R, Lund R, Hansen HV, Laenkholm AV, Tarin D, Jensen ON, et al. Metastasis-related plasma membrane proteins of human breast cancer cells identified by comparative quantitative mass spectrometry. Mol Cell Proteomics. 2009; 8: 1436-1449.
- Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer cell. 2005; 8: 393-406.
- Azimi A, Pernemalm M, Frostvik Stolt M, Hansson J, Lehtio J, Egyhazi Brage S, et al. Proteomics analysis of melanoma metastases: association between S100A13 expression and chemotherapy resistance. Br J Cancer. 2014; 110: 2489-2495.
- Deiss K, Kisker C, Lohse MJ, Lorenz K. Raf kinase inhibitor protein (RKIP) dimer formation controls its target switch from Raf1 to G protein-coupled receptor kinase (GRK) 2. The Journal of biological chemistry. 2012; 287: 23407-23417.
- Beach S, Tang H, Park S, Dhillon AS, Keller ET, Kolch W, et al. Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene. 2008; 27: 2243-2248.