Research Article
Expression of MicroRNA-224 in Cholangiocarcinoma and Its Clinicopathological Significance
Xiaolei WANG and Xiaofang LIU*
Department of Hepatobiliary Surgery, Affiliated Yantai Yuhuangding Hospital, Qingdao University Medical College, China
*Corresponding author: Xiaofang LIU, Department of Hepatobiliary Surgery, Affiliated Yantai Yuhuangding Hospital, Qingdao University Medical College, Yantai, P.R. China
Published: 06 Sep, 2018
Cite this article as: WANG X, LIU X. Expression of
MicroRNA-224 in Cholangiocarcinoma
and Its Clinicopathological Significance.
Clin Surg. 2018; 3: 2094.
Abstract
MicroRNAs (miRNAs) are widely involved in the regulation of various pathological and physiological
processes. One of the miRNAs, namely miR-224 expression has been found aberrant in a number of
human cancers; however, its expression and role in cholangiocarcinoma have not been studied. In
the present study, we investigated the difference of miR-224 expression in cholangiocarcinoma and
adjacent normal tissues and explored its clinicopathological significance. Methods: The expressions
of miR-224 in 30 cases of cholangiocarcinoma and its adjacent normal tissues were analyzed by RTqPCR.
The relationship between miR-224 and clinicopathological data was analyzed by statistical
analysis. Results: The expression levels of miR-224-5p in cholangiocarcinoma were significantly
higher than those in adjacent normal tissues (P<0.0001). The expression level of miR-224-5p in
cholangiocarcinoma was correlated with the pathophysiological behavior of tumor. That is, the
relative expression levels of miR-224-5p in patients with lymph node metastasis were significantly
higher than those in patients without lymph node metastasis (P<0.05);The relative expressions of
miR-224-5p in stage III and IV patients were significantly higher than those in stage I and II (P
<0.05). Conclusion: MicroRNA-224 is a potential tumor promoter in cholangiocarcinoma, which
may regulate the occurrence and development of cholangiocarcinoma and be used as a molecular
marker for the diagnosis and treatment of cholangiocarcinoma.
Cholangiocarcinoma is a highly invasive malignant tumor derived from cholangiocyte with a high
mortality rate in hepatobiliary surgery. In recent decades, its morbidity and mortality have been
increasing year by year [1]. Due to the insidious onset, rapid progression, difficulty in diagnosis,
insensitivity to conventional radiotherapy and chemotherapy, low resection rate, and high recurrence
rate, most patients with cholangiocarcinoma were diagnosed at an advanced stage and had lost
the opportunity for radical surgery [2]. Therefore, there is an urgent need to find a new molecular
marker that can evaluate the progression of cholangiocarcinoma, the prognosis of patients and the
possibility of targeted therapy to improve the quality of life or postoperative survival rate.
MicroRNA (miRNA) is a non-coding short RNA with a size of about 21-25 nucleotides. It is widely
involved in the regulation of various pathological and physiological processes, plays an important
role in cell proliferation, apoptosis, differentiation, and is closely related to the occurrence and
progression of tumors [3]. In recent years, a number of studies have shown that a variety of microRNAs
are expressed abnormally in cholangiocarcinoma and act as oncogenes or anti-oncogenes [4].
As a subtype of microRNA, miR-224 has been found to have abnormal expression in a variety of
malignant tumors and involved in the occurrence or progression of tumors [5,6]. However, there
is no report about its expression and its clinical pathological significance in cholangiocarcinoma.
Therefore, this study explored the differential expression of miR-224 in cholangiocarcinoma tissue
and adjacent tissues and its clinicopathological significance, in order to provide new ideas for the
prognosis of cholangiocarcinoma, and new targets for the precision medical treatment.
Materials and Methods
Patients
A total of 30 patients with distal bile duct cancer who underwent surgical treatment from
September 2015 to October 2017 in the Department of Hepatobiliary and Pancreatic Surgery
of Yantai Yuhuangding Hospital affiliated to Medical College of Qingdao University. The age
distribution of patients was 58-73 years old, with an average age of 65.30±4.324 years, and the
ratio of male to female 3:2. None of the selected patients had received previous radiotherapy or
chemotherapy.
All of them and their families had signed informed consent before
surgery and agreed to use their postoperative tissue specimens for this
study which was approved by the Hospital Medical Ethics Committee.
General information
All patients were definitively diagnosed finally by paraffin
pathological Examination with complete clinical and pathological
data. Among the patients, the postoperative pathology showed that
there were 10 cases of moderately or poorly differentiated and 20
cases of well-differentiated, including 16 cases with positive labeled
lymph nodes and 14 cases with negative.
Pathological staging and surgical variables
All patients were staged according to the 8th edition of the distal
cholangiocarcinoma staging system proposed by the American
Joint Committee on Cancer (AJCC):stage 0 is TisN0M0, stage I is
T1N0M0, stage II is T1N1M0, T2N0M0, T2N1M0, T3N0-1M0, stage III is
T1-3N2M0, T4NXM0, stage IV is TXNXM1 (Tis indicates carcinoma in
situ. T1 refers to the depth of tumor invasion is less than 5mm. T2
and T3 respectively indicate that the invasion depth is 5-12 mm and
greater than 12 mm.T4 indicates that the tumor invades celiac artery
trunk, superior mesenteric artery, and/or common hepatic artery;
N0 indicatesthat no regional lymph nodes were found to metastasize.,
N1 and N2 refer to 1-3, greater than or equal to 4 regional lymph node
metastases, respectively; M0 means no distant metastasis, M1 means
distant metastasis).
Among the 30 patients, there were 21 patients in stage I and II, and
9 patients in stage III and IV. Among them, 23 patients underwent
conventional pancreas to duodenectomy, a patient underwent
pylorus-preserved pancreas to duodenectomy, 2 patients underwent
biliary-jejunostomy, 3 patients underwent palliative tumor local
excision plus biliary enter story, a patient underwent laparotomy and
T-tube drainage.
Experimental reagents or Chemicals
Trizol solution was obtained from Shanghai Pufei Biotech,
reverse transcription primers and microRNA PCR primers were from
Guangzhou Ruibo Biotech, and reverse transcriptase kit was from
Promega Company of the United States.
Main instruments
F6/10 ultra-speed homogenizer form FLUKO Company,
Nanodrop2000/2000C spectrophotometer from Thermo Corporation,
LightCycler480 Real time PCR instrument form Swiss Roche.
Total RNA extraction
The tissue samples to be ground were taken out and cut to a size
of about 3mm × 3mm × 3mm on dry ice with a sterile blade. Place the
samples in EP tubes containing 1 mL of Trizol Lysis. After grinding,
centrifugation was performed for 3 min. The supernatant was pipetted
into new EP tubes. 200μL of chloroform was added to each tube, and
the EP tubes were inverted by hand for 15s, and left standing at room
temperature for 10 min. After 10 min, the mixture was centrifuged for
15 min, the supernatant was removed to new EP tubes, equal volume
of pre-cooled isopropanol was added, and the mixture was stand at
4ºC for 10 min. The supernatant was removed after centrifugation.
1mL 75% ethanol was used to wash the precipitate, the mixture was
centrifuged for 5min, after discarding most of the supernatant, and
the mixture was centrifuged again for 5min. The precipitation was
dried at room temperature after removing the supernatant. When the
RNA precipitation is substantially transparent, RNase-free water was
added until is completely dissolved, the concentration and quality
of the extracted RNA were determined by spectrophotometer. The
extracted total RNA was stored at -80ºC.
Reverse transcription to obtain cDNA
200 μl of RNase-free water was added per 1 nmol of the primer,
vortexed and fully dissolved, and then centrifuged instantaneously to
prepare the primer storage solution which having a final concentration
of 5 μm. 1 μL of 5 μm RT primer storage solution and add 79 μL of
RNase-free water were added to prepare 62.5 nM RT primer working
solution. PCR primers were used at a concentration of 5 μm. Then, 2
μL of reverse transcription primer, 2.0 μg of total RNA and RNase-
Free water were added into the PCR tubule to 11 μL. After mixing
and centrifuging, the mixture was placed at 70ºC for 10 min. Then the
mixture was placed in an ice-water mixture immediately to anneal
the reverse transcription primer and template. The above mixture
was mixed evenly in an ice bath to prepare a reaction system of
25μL. The above system was reacted in a 42ºC water bath for 1 h after
centrifuging briefly. Then the RT enzyme was inactivated in a water
bath at 70ºC for 10 min, and the obtained RT product-cDNA was
stored at -20ºC.
RT-qPCR detection
The reaction system (12 μL systems) was configured in the
following proportion: 6.0 μL of SYBR premix ex taq, 0.5 μL of upstream
primer, 0.5 μL of downstream primer, 1.0 μL of template, 4.0 μL of
RNase-Free water. In addition, RNA PCR reaction system(12 μL)
included 6.0μL of SYBR premix ex taq, 0.3 μL of primer mix, 0.6 μL
of template, 5.1μL of RNase-Free water. Performing Real-Time PCR
and making melting curves. The primer sequences were as follows:
MiR-224-5p RT Primer Sequence:
5′-C T C A A C T G G T G T C G T G G A G T C G G C A A T T
C A G T T G A G A A C G G A A C-3′,
Upstream primer sequence:
5′-ACACTCCAGCTGGGCAAGTCACTAGTGGTTCCGTT-3′,
Downstream primer sequence: 5′ -TGGTGTCGTGGAGTCG-3′;
U6: Upstream primer sequence: 5′-CTCGCTTCGGCAGCACA-3′,
Downstream primer sequence:
5′-AACGCTTCACGAATTTGCGT-3′.
U6 was used as an internal reference to normalize the miRNA
levels. The fold changes in gene expression of miRNA-224 were
calculated using the Livak 2–ΔCt method, where Ct is the crossing
threshold value and 2–ΔCt is the fold change in gene expression relative
to a reference gene.
Statistical method
All data are presented as mean ± standard deviation. Statistical
analyses were performed using Graph Pad Prism software, version
6. Differences between the groups were analyzed by Chi-Square test.
P<0.05 was considered statistically significant.
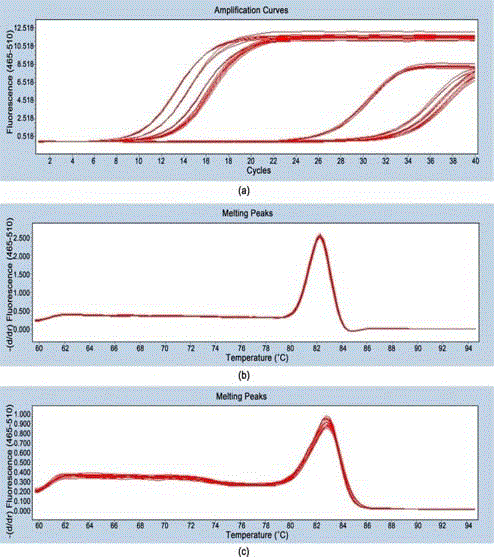
Figure 1
Figure 1
Detection of micro-224-5p and U6 primers.
Note: A, amplification curve of microRNA-224-5p and internal reference U6;
B, dissolution curve of U6; C, dissolution curve of microRNA-224-5p.
Figure 2
Result
Detection of miR-224-5p and internal reference U6 primers.
The amplification curves of miR-224-5P and U6 in some
cholangiocarcinoma tissues were constructed (Figure 1A). The
dissolution curves of miR-224-5P and U6 (Figure 1B,1C) are all
single-peak which suggested that the products of miR-224-5P and U6
are both single.
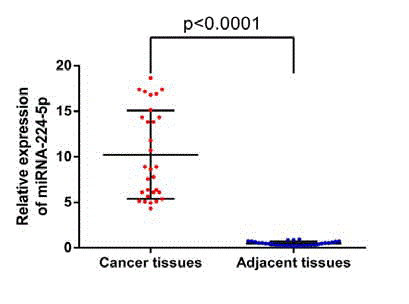
Expression of miR-224-5p in cholangiocarcinoma tissues
and adjacent tissues. The gene expression levels of miR-224 were
significantly higher in the human cholangiocarcinoma samples (10.23
± 0.8861) compared with the adjacent non-tumorous tissue samples
(0.4734 ± 0.04018), as determined by RT-qPCR assay. P<0.0001,
suggesting that the difference between the two is statistically
significant (Figure 2), (expanding 2–ΔCt by 10^6 times for drawing).
The relationship between the expression of miR-224-5p and the
clinicopathological features in patients with cholangiocarcinoma.
According to RT-qPCR, the relationship between the expression
of miR-224-5p and the clinicopathological features (age, sex,
differentiation, lymph node metastasis, pathological stage) was
analyzed. There was a significant correlation between the expression
level of miR-224-5p and lymph node metastasis in patients. The
relative expression of miR-224-5p in cancer tissues with lymph node
metastasis was significantly higher than that without lymph node
metastasis (P<0.05). In addition, the expression level of miR-224-
5p was found to be closely correlated with the pathological stage
of patients with cholangiocarcinoma. The relative expression level
of miR-224-5p in stage III and stage IV patients was significantly
higher than that in stages I and II (P <0.05). There was no significant
correlation between the relative expression of miR-224-5p and
clinicopathological factors such as age, sex and tumor differentiation
(P>0.05). (Table 1), (expanding 2–ΔCt by 10^6 times for data analysis).
Table 1
Table 1
Comparison of miR-224-5p Expression Levels in Cancerous Tissues of Patients with Cholangiocarcinoma with Different Clinicopathological Features.
Discussion
In recent years, the research of microRNAs in the field of
oncology has become more and more extensive. MiRNAs bind to
the 3' end of target mRNA by base-pairing principle and participate
in post-transcriptional gene regulation. Its expression errors are
closely related to the pathogenesis of many malignant tumors, and
have a wide range of biological effects [7]. There are characteristic
microRNA expression profiles in tumor tissues which are different
from normal tissues, and the expression changes of these microRNAs
can all be determined in the laboratory. Therefore, microRNAs may
become a new molecular marker for tumor diagnosis or disease
progression and a molecular target for judging tumor prognosis,
which will benefit the diagnosis and treatment of tumor [8].
Studies have shown that many miRNAs are found to be
abnormally expressed in tumors, in which miR-224 has received
much attention. As a member of the microRNA family, miR-224 has
been found to have significant abnormal expression and function as
oncogenes or anti-oncogenes in a variety of human tumors such as
liver cancer [9], gastric cancer [10], colorectal cancer [11], pancreatic
cancer [12], diffuse large B-cell lymphoma [13], prostate cancer [14],
breast cancer [15], cervical cancer [16], non-small cell lung cancer
[17] and so on. Determinating its expression changes may become
a new and important means for early diagnosis and prognosis of
malignant tumors.
In this study, we selected the surgically resected
cholangiocarcinoma tissue and adjacent non-tumorous tissues and
detected the relative expression levels of miR-224 by RT-qPCR. The
results showed that the expression level of miR-224 was significantly
up-regulated in cancer tissues compared with adjacent tissues,
suggesting that miR-224 plays a role as a potential oncogene in
cholangiocarcinoma.
Combined with the clinicopathological data of patients,
the relationship between the expression of miR-224 and
clinicopathological features of patients was analyzed by statistical
methods. The results showed that the expression level of miR-224
was not correlated with the age, gender and tumor differentiation of
patients. However, its level in patients with lymph node metastasis was
significantly higher than that without lymph node metastasis. And the
later the pathological stage in patients, the higher the expression level
of miR-224.The experimental results suggest that high expression
of miR-224 may have a negative effect on the prognosis of patients
with cholangiocarcinoma. Therefore, it is speculated that miR-224
may regulate the occurrence, progression, migration and invasion
of tumors, which may provide a theoretical basis and reference for
the prognosis of patients and a new target for precise treatment of
cholangiocarcinoma.
Of course, this study also has limitations such as fewer patient
cases and lack of follow-up. In addition, the precise value of miR-
224 in the prognosis of patients with cholangiocarcinoma cannot be
further evaluated.
At present, the research on microRNA-224 is not thorough
enough. This study laid the foundation for the study of the
regulation mechanism and signaling pathway of microRNA-224
and cholangiocarcinoma. The specific target genes involved in the
regulation of microRNA-224 and the detailed molecular mechanisms
involved in the biological function of cholangiocarcinoma require
further improvement or exploration.
Conclusion
The above results reveal that the expression levels of miR-224- 5p in cholangiocarcinoma were significantly higher than those in adjacent normal tissues. The expression level of miR-224-5p in cholangiocarcinoma was correlated with the pathophysiological behavior of tumor. MiR-224 may play a role in the process of occurrence, progression, invasion and migration of cholangiocarcinoma. These findings suggest that miR-224 is being assessed as a potential new target in the treatment and a novel biomarker for diagnosis and prognosis prediction of cholangiocarcinoma.
Acknowledgements
The work was supported by grants from Natural Science Foundation of Shandong Province, P.R. China (No.ZR2014HM052).
References
- Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168-79.
- Khan SA, Toledano MB, Taylor-Robinson SD. Epidemiology, riskfactors, and pathogenesis of cholangiocarcinoma. HPB (Oxford). 2008;10(2):77-82.
- Sayed D, Abdellatif M. Micrornas in development and diserse. Physiol Rev. 2011;91(3):827-87.
- Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1-20.
- Chen W, Fan XM, Mao L, Zhang JY, Li J, Wu JZ, et al. MicroRNA-224:as a potential target for miR-based therapy of cancer[J]. Tumour Biol. 2015;36(9):6645-52.
- Tang ZH, Tian XD, Wei MY. Updates and interpretations of the 8th edition of AJCC cancer staging system forbiliary tract carcinoma. Chinese Journal of Practical Surgery. 2017;37(3):248-54.
- Wang J, Sen S. MicroRNA functional network in pancreatic cancer: from boilogy to biomarker of disease. J Biosci. 2011;36(3):481-91.
- Budhu A, Ji J,Wang XW. The clinical potential of microRNAs. J Hematol Oncol. 2010;3:37.
- Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, et al. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59(2):505-17.
- Liu H, Li P, Li B, Sun P, Zhang J, Wang B, et al. RKIP suppresses gastric cancer cell proliferation and invasion and enhances apoptosis regulated by microRNA-224. Tumour Biol. 2014;35(10):10095-103.
- Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. Up-regulation of miR-224 promotes cancercell proliferation and invasion and p-redicts relapse of colorectal cancer. Cancer Cell Int. 2013;13(1):104.
- Zou YQ, Liu C, He WF. Effect and mechanism of antisense miR-224 on invasion and migration of pancreaticcarcinoma cell. Chin J Clin Lab Sci. 2014;32(3):212-7.
- Ni H, Wang X, Liu H, Tian F, Song G. Low expression of miRNA-224 predicts poor clinical outcome in diffuse la-rge B-cell lymphoma treated with RCHOP. Biomarkers. 2015;20(4):253-7.
- Wan Y, Zeng ZC, Xi M, Wan S, Hua W, Liu YL, et al. Dysregulated microRNA-224/apelin axis associated with aggressive progression and poor prognosis in patients with prostate cancer. Hum Pathol. 2015;46(2):295-303.
- Feng XB, Zhao L, Gao SG, Song X, Dong W, Zhao Y, et al. Increased fucosylation has a pivotal role in multidrug resistance of breast cancer cells through miR-224-3p targeting FUT4. Gene. 2016;578(2):232-41.
- Wang F, Shan S, Li YM, Zhu E, Yin L, Sun B. miR-224-3p inhibits autophagy in cervical cancer cells by targeting FIP200. Sci Rep. 2016;6:33229.
- Xu ZY, Zhou JL, Zhou J. expression and clinical significance of MicroRNA-224-5p expression in NSCLC. Practica lMedical Journal. 2016;32(21):3463-7.